Combining (joining/merging) data sets with dplyr
Cole Tanigawa-Lau
Functions covered:
Combining data sets
It is nearly always the case that your analyses will require data from a combination of at least two distinct data sets. The data sets you see in class and use for assignments have already been cleaned and merged for you. The analyses for your final projects, however, will likely require using variables from multiple data sources and combining them based on a variable the data sets share. This process is commonly known as merging in the social sciences and joining in database contexts. Here, I’ll introduce the types of dplyr joins that I have used most frequently. For a much more extensive demonstration of joins in dplyr, you can check out this vignette. You can also use this cheat sheet as a reference.
About the data
We’ll be working with the flood and campaign contributions data sets that we built in the aggregation lesson. These data sets all have a variable in common: zip code. Having a shared variable to link each data set will be crucial in telling R how the data should be joined.
library(dplyr)
contrib <- readRDS("data/zip_contrib.rds")
flood <- readRDS("data/zip_flood.rds")Checking the data
First, let’s take a look at the data we’ll be combining.
head(contrib)## # A tibble: 6 x 2
## zip contrib_amount
## <chr> <dbl>
## 1 07001 250
## 2 07002 4685
## 3 07003 22330
## 4 07004 46335
## 5 07005 33210
## 6 07006 157766head(flood)## # A tibble: 6 x 2
## zip flood_extent
## <chr> <dbl>
## 1 08310 0
## 2 08403 0.905
## 3 08205 0.503
## 4 08221 0.501
## 5 07430 NA
## 6 07650 0.156Before merging, it’s a good idea to check either the overlap or difference between each data set’s linking variable, (in this case, zip codes). Figuring out any problems at this point could save a good deal of time and frustration down the road. The functions setdiff() and intersect() are useful here.1
setdiff(contrib$zip, flood$zip)## [1] "07101" "07303" "07451" "07474" "07507" "07875" "07962" "07963"
## [9] "07978" "08213" "08218" "08250" "08362" "08504" "08543" "08739"
## [17] "08754" "08818"setdiff(flood$zip, contrib$zip)## [1] "08064" "08042" "08224" "08105" "08091" "08251" "08352" "08327"
## [9] "08320" "08066" "08061" "08328" "08828" "08733" "07505" "07439"
## [17] "08346" "08319" "07074" "08640" "08641" "08554" "08041" "08045"
## [25] "08049" "08110" "08230" "08311" "08314" "07108" "07878" "07870"
## [33] "07504" "07524" "08038" "08890" "07851" "07846" "08808" "08317"
## [41] "08562" "08073" "08104" "08030" "08007" "08248" "08323" "08316"
## [49] "07514" "07880" "08011" "08089" "08004" "08324" "07106" "08074"
## [57] "07077" "08810" "07420" "08821" "07822" "08344" "07606" "08031"
## [65] "08102" "08095" "08345" "08039" "07311" "07865" "08609" "08832"
## [73] "08722" "07502" "08215" "08340" "08240" "08019" "08246" "08318"
## [81] "08329" "08321" "08090" "07501" "08067" "07063" "08880" "07881"
## [89] "08350" "08103" "08084" "08029" "08348" "08063" "08097" "08629"
## [97] "07421" "07939" "07833" "08326" "08217" "07660" "07495" "08078"
## [105] "08083" "08353" "08093" "08014" "08027" "07064" "07721" "07703"
## [113] "08555" "07857" "07970" "07513" "07522" "08079" "08001" "08072"
## [121] "07206"zip_inter <- intersect(flood$zip, contrib$zip)
length(zip_inter)/length(flood$zip)## [1] 0.7966387There are many zip codes included in the flood data set that are missing in the contributions data set. In some cases, this could indicate a problem in the data gathering or cleaning stages. Here, however, it makes sense: not all zip codes contributed to Gov. Christie’s reelection campaign. It looks like almost 80% of the zip codes will merge properly, so let’s go ahead and check each data set for any duplicated zip codes.
any(duplicated(contrib$zip))## [1] FALSEany(duplicated(flood$zip))## [1] FALSEJoining
R’s base function for joining data is merge(), which offers many arguments to control the join. In dplyr, additional functionality is offered through multiple joining functions. We will cover the most common type of join, in which you are combining two data sets. To learn about subsetting one data set based matching values in another, see the section on filtering joins in the vignette linked above.
As we go through the different types of joins, it might help to refer to this diagram made by Hiroaki Yutani: 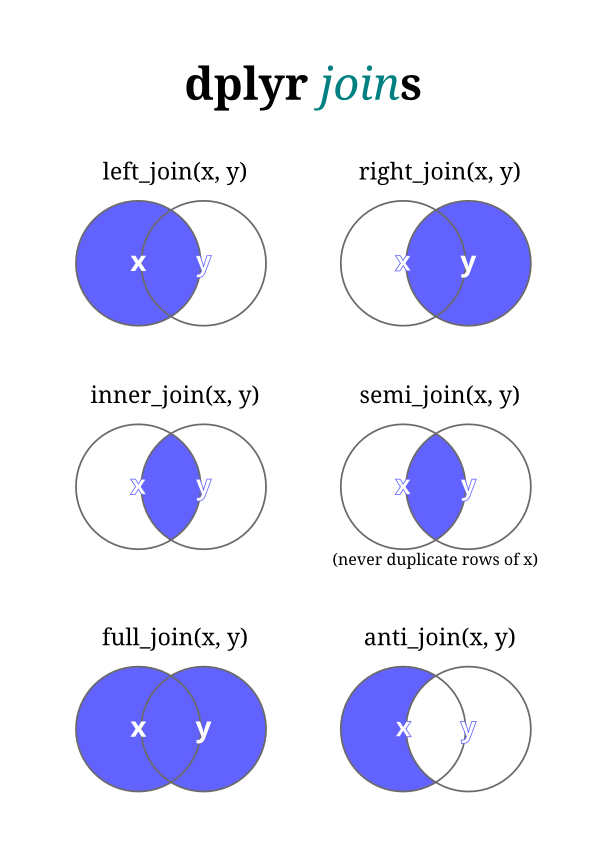
inner_join()
The inner_join() function returns a data frame containing only observations with a match in both data sets. In other words, our result will contain only rows with zip codes that are in both contrib and flood. The inner_join() function is equivalent to using base::merge() with the default parameters.
Since the only column we want to use as a reference has the same name in both data sets, we don’t need to specify a by argument for any of the dplyr joins, but I’ll do so here for clarity and to avoid a message from dplyr.
data_ij <- inner_join(contrib, flood, by = "zip")
# Recall that we assigned zip_inter <- intersect(contrib$zip, flood$zip)
all(data_ij$zip %in% zip_inter)## [1] TRUEnrow(data_ij)## [1] 474As you can see, we lost many observations in data_ij, including all of the zip codes that did not make any campaign contributions. That doesn’t make much sense, since these amounts should actually be zero. For our purposes here, we’ll want to use a different joining function.
left_join() and right_join()
The left_join() and right_join() functions return a data frame containing all observations in one data frame and the matching observations from the other.
data_lj <- left_join(contrib, flood, by = "zip")
nrow(contrib) == nrow(data_lj)## [1] TRUEThe left_join() function takes the data frame on the “left” (i.e., contrib, or the first one passed to left_join()) and adds on the matching columns from the “right” data frame. The result is a data frame with the same number of rows as contrib. We can see that iforn zip codes not contained in flood_extent, there are NA values in data_lj.
filter(data_lj,
is.na(flood_extent)) %>%
head()## # A tibble: 6 x 3
## zip contrib_amount flood_extent
## <chr> <dbl> <dbl>
## 1 07101 20 NA
## 2 07303 1000 NA
## 3 07401 18425 NA
## 4 07403 1000 NA
## 5 07416 1375 NA
## 6 07418 400 NAThe right_join() function is the mirror image of left_join(). It’s as if you switched the x and y arguments in left_join().
data_rj <- right_join(contrib, flood, by = "zip")
nrow(flood) == nrow(data_rj)## [1] TRUEsetdiff(data_rj, left_join(flood, contrib, by = "zip"))## # A tibble: 0 x 3
## # ... with 3 variables: zip <chr>, flood_extent <dbl>,
## # contrib_amount <dbl>full_join()
The full_join() function returns a data frame containing all observations from both data frames. This is useful for when you don’t want to exclude any observations simply due to missingness.
data_fj <- full_join(contrib, flood, by = "zip")
setdiff(data_fj$zip, c(contrib$zip, flood$zip))## character(0)Duplicate matches
So far, we’ve covered fairly basic joins with exact one-to-one matches. Very commonly, though, you’ll be merging data at different levels or with some duplicate values in the linking variable. We’ll use the individual-level campaign contributions data set from our aggregation lesson as an example.
indv_contrib <- readRDS("data/indv_contrib.rds")
indv_data <- full_join(indv_contrib, flood, "zip")There are many more observations here for each zip code, so let’s look at only one zip code to see what’s going on. When joining, we took the flood_extent value for zip code 08008 and matched it to all observations in indv_contrib with the same zip code. Because flood_extent is specific only to zip codes and not to the individual donors that live in these areas, indv_data$flood_extent has many duplicate values.
# Single observation in flood data
filter(flood, zip == "08008")## # A tibble: 1 x 2
## zip flood_extent
## <chr> <dbl>
## 1 08008 0.565# That value is entered in the "flood_extent" column for all individuals
filter(indv_data, zip == "08008")[1:10, ]## id name amount date city
## 1 7234317 BARRON, ART R 3400 2009-09-21 BEACH HAVEN
## 2 7234621 MANCINI, JOSEPH H 3400 2009-09-22 BEACH HAVEN TERRACE
## 3 7234612 BARRON, JOAN C 1500 2009-09-22 BEACH HAVEN
## 4 7235259 RUSSO, CAROLA A 1000 2009-08-21 LONG BEACH TOWNSHIP
## 5 7233166 MUROFF, ROBERT 500 2009-09-28 LONG BEACH TOWNSHIP
## 6 7233178 SHACKLETON, RICHARD J 500 2009-09-28 SHIP BOTTOM
## 7 2188126 BATTISTA, MICHAEL 500 2009-09-28 BEACH HAVEN
## 8 5215201 HUTSON, WILLIAM E 500 2009-09-28 LONG BEACH TOWNSHIP
## 9 981749 CLEARY, DENNIS J 500 2009-09-28 BEACH HAVEN
## 10 7233296 LEONETTI, JIM 500 2009-09-28 LONG BEACH TOWNSHIP
## zip flood_extent
## 1 08008 0.5646336
## 2 08008 0.5646336
## 3 08008 0.5646336
## 4 08008 0.5646336
## 5 08008 0.5646336
## 6 08008 0.5646336
## 7 08008 0.5646336
## 8 08008 0.5646336
## 9 08008 0.5646336
## 10 08008 0.5646336Loading the
dplyrpackage imports set functions likeintersect()andsetdiff(), making them available for use on data frames, but they’ll still work on vectors as well.↩